3D Microfluidic Cancer Cell Chemoinvasion Model
Although cancer metastasis is a leading cause of cancer-related deaths in humans, the process has been largely unknown, primarily due to its dynamic nature and the complex interaction of tumor cells with their environments. Microfluidic devices offer an opportunity to study this dynamic process on physiologically relevant time and length scales. We have used an agarose-based three-channel device to build a 3D cell culture in a collagen matrix and to generate a stable gradient of candidate chemical signal molecules. With different cytokines and growth factors, we found that a malignant breast tumor cell line, MDA-MB-231, shows chemotaxis towards CXCL12 and CCL21 gradients, while it presents chemokinesis under EGF and CCL19 gradients. In addition, there were high plasticity and heterogeneity in the tumor population, in terms of their morphologies and motility patterns. The probability distribution of the displacement of MDA-MB-231 cells showed a Gaussian core with thicker tails, implying that the dissemination strategy may be different from the random walk theory. This approach will eventually lead to an insight into the strategy related to cancer dissemination.
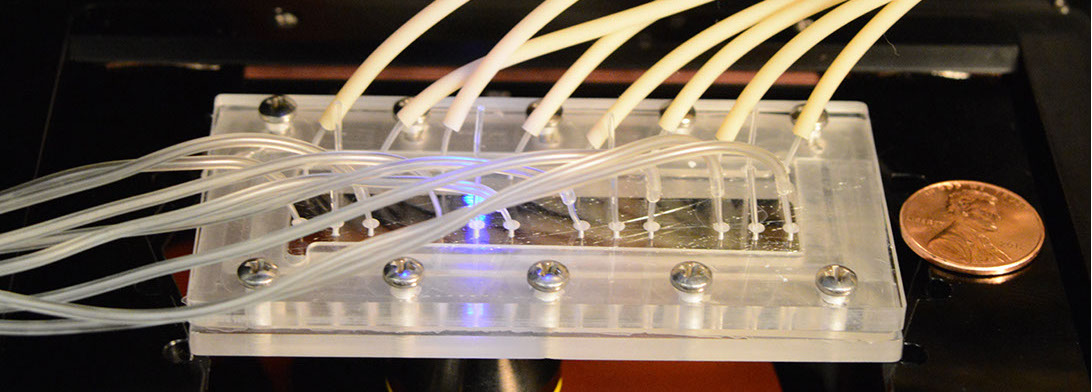
The set up of chemoinvasion experiment to generate chemokine gradients using microfluidics.
Publications:
- Beum Jun Kim, Pimkhuan Hannanta-anan, Michelle Chau, Yoon Soo Kim, Melody A. Swartz, and Mingming Wu, Cooperative roles of SDF-1a and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model, PLoS ONE 8(7): e68422. doi:10.1371/journal.pone.0068422 (2013).
- Beum Jun Kim and Mingming Wu, Microfluidics for mammalian cell chemotaxis, invited review, Annals of Biomedical Engineering, 40, 1316-1327 (2012).
Site created and maintained by Young Joon Suh

