Chemotaxis
Chemotaxis is a process where cells move up/down in response to a chemical gradient. It is one of the main routes that cells use to communicate with each other. Here, we investigated chemotaxis process of various cell types, bacteria, sperm, immune and cancer cells. We found that these cellular chemosensitivity is all governed by the ligand biding kinetics, however, the level of the sensitivity varies dramatically depending on species.

Dendritic Cell Chemotaxis -- How do dendritic cells sense and move? (Rikki Haessler and Eugene Kalinin)
Immune cells are your first line of defense when you get a pathogen attack. How do immune cells coordinate and migrate to the site of injury? In this work, we illustrated that dendritic cells are directed to lymph nodes by chemokine gradients. A dendritic cell compares the ligand receptor binding sites at the front and back of the cell, and then make a move decision. For more details, please read Haessler et al., Biomed Microdevice, 2009 and PNAS, 2011.
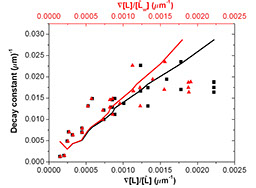
Bacterial Chemotaxis -- Do E.coli sense chemical gradients at a logarithmic scale ?(Eugene Kalinin)
We use Escherichia coli (E.coli) as a model system to explore underlying mechanism that cells use to navigate in a chemical gradient. The graph here shows that chemosensitivity is linearly related to the log of the chemoattractant concentration gradient. You can find more information in Kalinin et al. Biophys J., 2009.
Who is the winner? – Probing the potency of multiple chemoattractant Escherichia coli (Eugene Kalinin)
By flowing the two side channels of our microchemotaxis device with different chemicals one can establish and maintain two independent gradients of different chemicals in the center channel. Chemotactic responses of E. coli to these "competing concentration gradients" provide clues on the comparative roles of the receptors in the cells’ chemotactic behavior. You can find more information in Kalinin et al., Biophys J. 2010.
Publications:
1.Haixin Chang*, Beum Jun Kim*, Yoon Soo Kim, Susan Suarez, Mingming Wu, Differential migration pattern of sea urchin and mouse sperm revealed by a microfluidic chemotaxis device, PlosOne, DOI: 10.1371/journal.pone.0060587 (2013)
2.Ulrike Haessler, Marco Pisano, Mingming Wu and Melody A. Swartz, Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to CCL21 and CCL19, PNAS, 108 (14) 5614-5619 (2011).
3.Kalinin, Y., Neumann, S., Soujik, V., & Mingming Wu. Responses of Escherichia coli Bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio, Journal of Bacteriology, 192, 1796-1800 (2010).
4. Haessler, U., Y. Kalinin, M.A. Swartz, and Mingming Wu, An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies. Biomedical Microdevices, 11(4): p. 827-35, (2009).
5. Yevgeniy Kalinin, Lili Jiang, Yuhai Tu, and Mingming Wu, Logarithmic sensing in Escherichia coli bacterial chemotaxis, Biophysical J, 96(6): p. 2439-48, (2009).
6. Shing-Yi Cheng, Steve H. Heilman, Max Wassweman, Shivaun Archer, Michael L. Shuler and Mingming Wu, A hydrogel-based microfluidic device for the studies of directed cell migration. Lab-on-a-chip, 7: p. 763-769, (2007).
7. JinPian Diao, Lincohn Young, Sue Kim, Elizabeth A. Fogarty, Steve M. Heilman, Peng Zhou, Mike L. Shuler, Mingming Wu, DeLisa,M.P, A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab on a Chip, 6: p. 381-388l, (2006).
Site created and maintained by Young Joon Suh

