Statistical measures of swimming E.coli

E. coli is a single cell organism with a cigar shape, the length of a bacterium is typically 1-3 micrometer and diameter ~ 1 micronmeter. Each bacterium has its own set of variable speed motor with propellers (filagella) enabling it to swim at speeds approaching 40 um/second. It is also equipped with its own sensing and signaling networks providing it with the ability to swim smartly (as opposed to randomly) in response to neighboring cells and/or chemical cues around it.
The Reynolds number of an E.coli cell is typically on the order of 10-4, making the world of E.coli very different from you and I. Inertia does not play any roles in its life. As a result, a bacterium can change from a run to a tumble or vice versa very abruptly.
It is known that bacteria's ability to communicate plays an important role in their ability to disperse, to lauch an attack to a host cell, or to form biofilms. We are interested in studying bacteria's collective dynamics when they are subjected to different chemical and nutrient environment. Our approach is to track positions of multiple swimming bacteria both in time and space. The tracking data provides a vast amount of information about swimming bacteria both macroscopically (diffusion coefficient) and microscopically (flagella motor power, cell-cell interaction). On the left is a movie of swimming bacteria (RP437, gift from Prof. Parkinson at University of Utah)
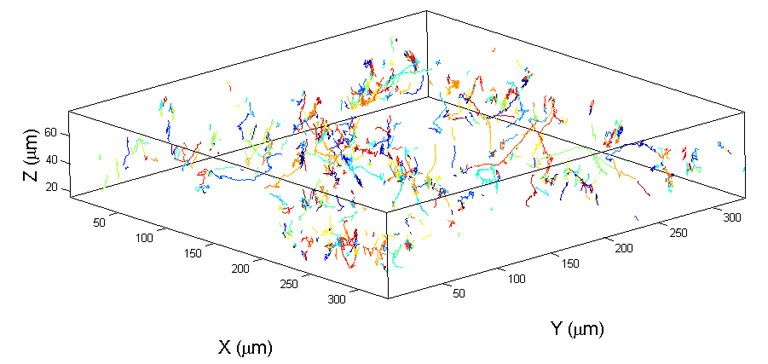
630 bacterial tracks of wildtype RP437.
Publications:
- Qian Liao, John Roberts, Matthew P. DeLisa, and Mingming Wu, Statistical measures of swimming Escherichia coli, submitted to PRL, (2005).
- Mingming Wu, John Roberts, Sue Kim, Donald L. Koch and Matthew P. DeLisa, Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique, Applied and Environmental Microbiology, accepted, (2006).
Site created and maintained by Young Joon Suh

